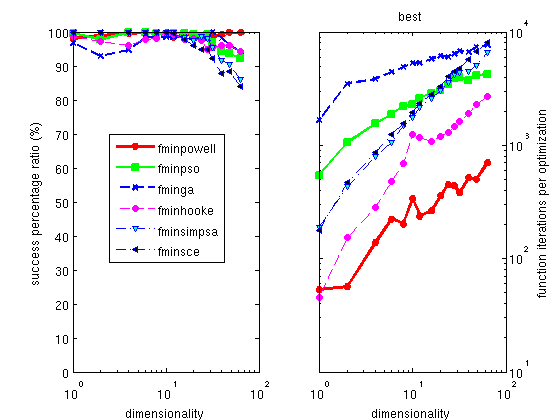
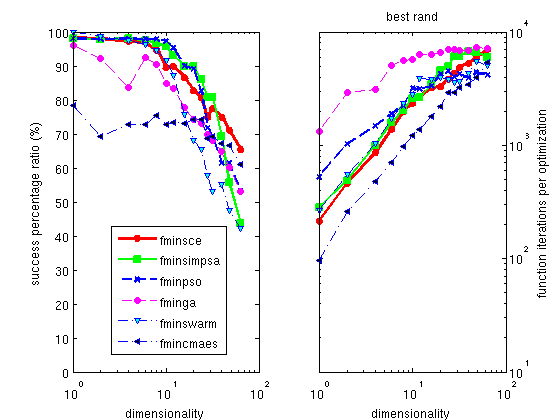
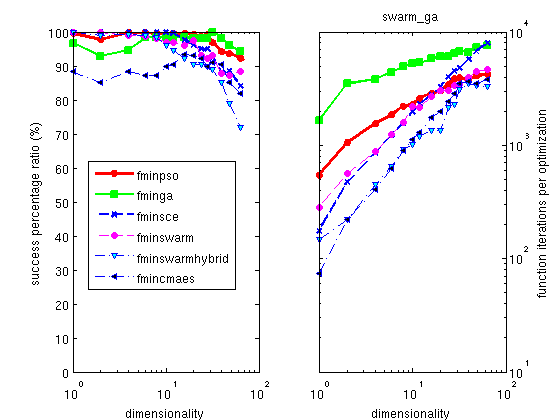
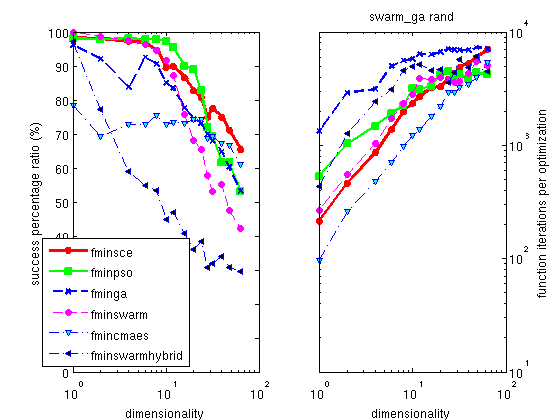
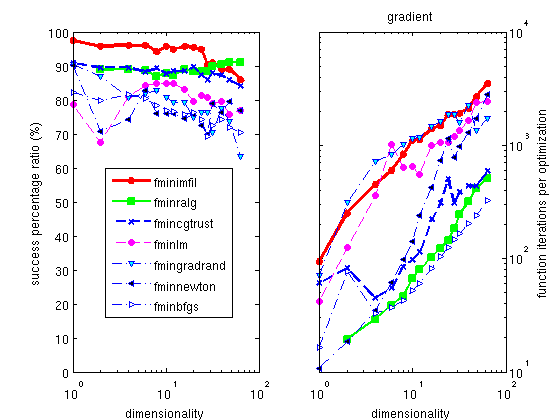
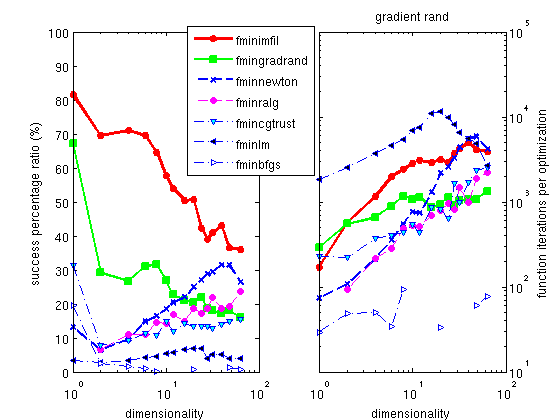
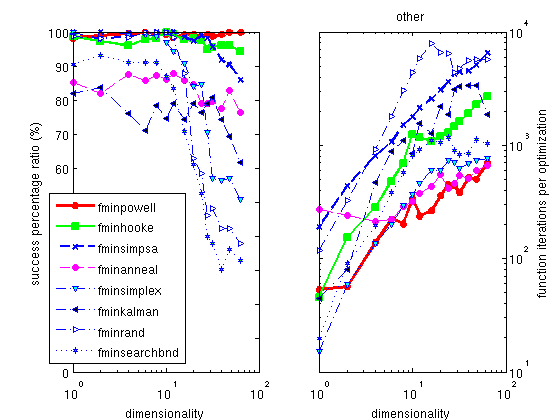
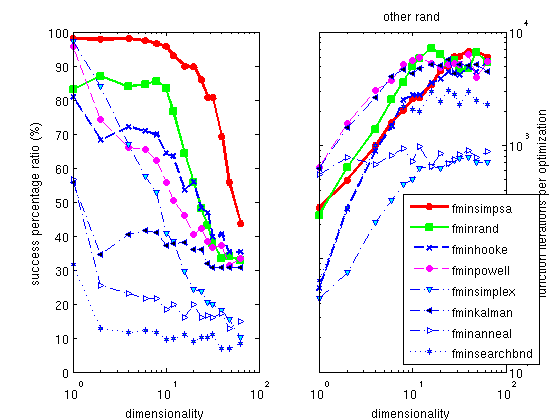
The Hooke optimizer
fminhooke
is one of the most efficient methods for all continuous problem,
and among the fastest solving time.
The Powell optimizer
fminpowell is a well known, simple
and efficient method, which partly works on noisy problems. Its
version with the Coggins line search is the fastest of all the
optimizers proposed here. The use of the Golden rule line search
slightly improves its success ratio, but dramatically increases
the execution time.
The default optimizer shipped with Matlab is the
fminsearch one, which is a
pure simplex implementation. We do not recommend this method, as
all others are better in success ratio, some being as fast. A
better alternative is a mix of a simplex with a simulated
annealing process, implemented as
fminsimpsa.
Model parameter constraints and
additional arguments
In many cases, the model parameters are to be constrained. Some
optimization specialists call these
restraints, that is parameter values
constraints. This includes some fixed values, or bounds within
which parameters should be restricted. This is specified to the
optimizer method by mean of a 4th input argument
constraints
:
>> [parameters,criteria,message,output]= fminimfil(model, initial_parameters, options, constraints)
In short, the constraints is a structure with the following
members, which should all have the same length as the model
parameter vector:
- constraints.fixed: 1
for fixed parameter, 0 for free parameters
- constraints.min: the
minimum
value for each parameter.
-Inf is supported. NaN can be used.
- constraints.max: the
maximum
value for each parameter.
+Inf is supported. NaN can be used.
- constraints.steps: the
maximum
change between iterations for each parameter. +Inf is supported.
- constraints.eval: any other expression to
evaluate, returning a modified parameter vector 'p'
All these constraints may be used simultaneously.
The
constraints input
argument can also be entered as a character string, like the
input
parameters and
options :
constraints='min=[0
0 0 0]; max=[1 10 3 0.1]; eval=p(4)=0';
Fixed
parameters
To fix some of the model parameters to their starting value, you
just need to define
constraints
as a vector with 0 for free parameters, and 1 for fixed
parameters, e.g. :
>> p=fminimfil(objective, starting, options, [ 1 0 0 0 ])
will fix the first model parameter. A similar behavior is
obtained when setting constraints as a structure with a
fixed member :
>> constraints.fixed = [ 1 0 0 0 ];
The
constraints vector
should have the same length as the model parameter vector.
Parameters
varying within limits
If one needs to restrict the exploration range of parameters, it
is possible to define the lower and upper bounds of the model
parameters. This can be done by setting the 4th argument to the
lower bounds
lb, and
the 5th to the upper
ub,
e.g. :
>> p=fminimfil(objective, starting, options, [ 0.5 0.8 0 0 ], [ 1 1.2 1 1 ])
A similar behavior is obtained by setting constraints as a
structure with members
min
and
max :
>> constraints.min = [ 0.5 0.8 0 0 ];
>> constraints.max = [ 1 1.2 1 1 ];
The
constraints
vectors should have the same length as the model parameter
vector, and
NaN values
can be used not to apply min/max constraints on specified
parameters.
Limiting
parameter change
Last, it is possible to restrict the change rate of parameters
by assigning the
constraints.steps
field to a vector. Each non-zero value then specifies the
absolute change that the corresponding parameter can vary
between two optimizer iterations.
NaN values can be used not to apply step
constraints on specified parameter.
Other
constraints/restraints
The constraints.eval
member can be used to specify any other constraint/restraint
by mean of
- either an expression making use of 'p', 'constraints','
and 'options', and returning the modified 'p' values (this
expression is evaluated) ;
- or a function handle of 'p', returning modified 'p'
values.
For instance one could use constraints.eval='p(3)=p(1);'.
Additional
arguments to the objective
If the objective function requires additional arguments (which
are not part of the optimization, and are kept constant)
objective(p, additional_arguments)
the syntax for the optimization is e.g.
>> p=fminsce(objective, [ 0.5 1 0.01 0 ],options, constraints, additional_arguments);
that is additional arguments are specified as 5th, 6th...
arguments. In this case, would you need to set optimizer
configuration or restraints/constraints, we recommend to give
'options' and 'constraints' arguments 3-4th as structure, as
explained
above.
The type of the additional arguments can be anything: integer,
float, logical, string, structure, cell...
For instance, we define a modified Rosenbrock 2 parameters
objective which prints a star each time it is evaluated:
>> banana = @(x, cte) 100*(x(2)-x(1)^2)^2+(1-x(1))^2 + 0*fprintf(1,'%s', cte);
>> p=fminsce(banana, [ 0.5 1 0.01 0 ],'', '', '*');
.......................
ans =
0.1592 0