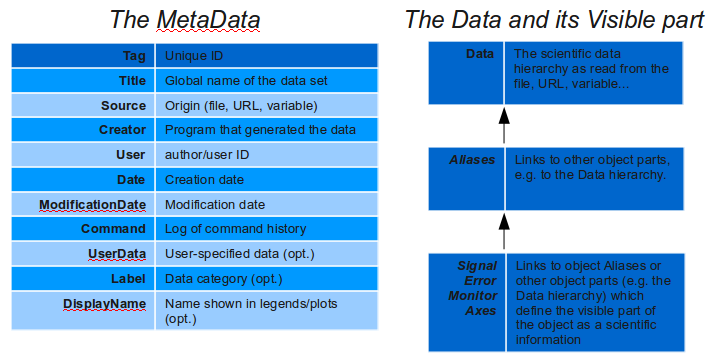
The iData object internal internal structure.
>> a = iData([ ifitpath 'Data/ILL_IN6.dat']) % imports a file into an object
a = iData object:
[Tag] [Dimension] [Title] [Last command] [Label]
tp460929 [257 512] 'File ILL_IN6.dat ILL Data (n...' tp460929=load(iData,...
>> a = iData(rand(10))These objects, which internal structure can be obtained with the struct(a) method, hold a number of important items:
>> a = iData(x,y,signal) % give the axes first, then the Signal
>> a = iData(gauss, 5, -5:5) % a gaussian with width=5, evaluated along axis -5:5
>> a = iData([ ifitpath 'Data/ILL_IN6.dat' ]); % converts a file name into an iData objectshowing the object unique ID, its dimensions, title, the last command which produced the object, and an optional label (here empty). The same output is given by the display(a) command.
>> a
ans = iData object:
[Tag] [Dimension] [Title] [Last command] [Label]
tp481575 [257 512] 'File ILL_IN6.dat ILL Data (n...' tp481575=load(iData,...
>> get(a)
>> disp(a)
a = iData 2D object of size [257 512]: |
>> a.Signal % returns the Signal valueA single field can be accessed in an object or an array of objects, by means of the get function (just like for the Matlab graphics objects):
>> a{0} % returns the Signal/Monitor value
>> a{1} % request the 1st axis values
>> a=load(iData, [ ifitpath 'Data/*.scn']) % returns an array of iData objectswhere this latter is equivalent to accessing
>> get(a,'Title') % returns the Title of all objects in the array
>> get(a(1),'Title') % returns the Title from the first object
>> a(1).TitleField names are case insensitive, that is b.title will also work.
>> b=a(1)
>> b.Title
>> get(a,'Data.VARIA.A1') % returns all A1 values from the array of objectsAccessing subfields requires a case sensitive name match, that is a a(3).data works (single field accessed), but a(3).data.varia.a1 does not (requires fully qualified path to the data).
>> a(2).Data.VARIA.A1 % same but only for the second array element
>> a(3).Data % returns the complete Data structure from the 3rd object in the array
>> a=load(iData, [ ifitpath 'Data/sv1850.scn'])Some data file formats provide additional 'attributes' to each numeric content (e.g. NeXus/HDF/NetCDF/CDF. See Loaders). You can access these associated attributes with the fileattrib method:
>> [match, field]=findstr(a,'TAS');% searches matches within object contents
match =
'UPt3;File sv1850.scn ILL TAS Data'
[1x105 char]
[1x169 char]
'ETAS= '
field =
'Title'
'Creator'
'Data.Attributes.AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA'
'Data.Attributes.PARAM.ETAS'
>> findfield(a,'TAS') % searches matches within object members/aliases/fields
ans =
'Data.PARAM.ETAS'
'Data.Attributes.PARAM.ETAS'
>> fileattrib(a, 'Data.Attributes.PARAM.ETAS')The findobj method can be used to inquire all iData objects available in the workspace:
ans =
ETAS=
>> a=iData([ ifitpath 'Data/IRS21360_graphite002_ipg.nxs' ]);
>> getalias(a,'Signal')
ans =
Data.mantid_workspace_1.workspace.values
>> fileattrib(a,'Signal')
ans =
signal: 1
axes: 'axis1,axis2'
units: ''
unit_label: ''
>> a = findobj(iData);It is also possible to plot the object, and we direct the reader to the Plotting page.
>> a = findobj(iData, 'Property', 'Value'); % searches for objects that match a Property/Value pair
>> a = plot(a);
>> a = edit(iData);
>> a=load(iData, [ ifitpath 'Data/*.scn']) % returns an array of iData objectsThe new assignments apply on the object itself when the method call has no output argument, but can additionally be copied to an other variable:
>> set(a,'Data.VARIA.A1',40); % modify the whole array Data content
>> set(a(1),'Data.VARIA.A1',40); % modify a single Data content
>> b = set(a(1),'Data.VARIA.A1',40); % modify the Data content and send back the modified objectThe set assignment may also be used with arrays of iData objects.
>> a=load(iData, [ ifitpath 'Data/sv1850.scn'])We shall see below that there is a way to extend an iData object, that is add some new information, and to create links within the object.
>> a.Data.VARIA.A1
ans =
40
>> a.Data.VARIA.A1 = 41 % returns the modified object
>> a=load(iData, [ ifitpath 'Data/sv1850.scn'])
>> a.Title
ans =
UPt3;File sv1850.scn ILL TAS Data
>> a.Label = 'TAS'These can be searched using the findfield, and findstr methods, see above, or findobj(iData,'Label','TAS').
a = iData object:
[Tag] [Dimension] [Title] [Last command] [Label]
tp120236 [15 1] 'UPt3;File sv1850.scn ILL TAS...' tp120236=set(tp12023... TAS
>> a=load(iData, [ ifitpath 'Data/sv1850.scn'])In this latter case, that is when Aliases are defined as numerics (scalar, vector, matrix), this is strictly equivalent to
>> setalias(a,'NewField',42)
>> a.NewField = 42The mechanism seen here works for numerical fields, but will fail if you decide to store structures and cell arrays into an Alias. Then, we recommend to define a new member in the Data part of the object, and then point to it as a link, as explained below.
>> set(a,'NewField',42)
>> setalias(a,'NewField',42,'Answer to the Ultimate Question of Life, the Universe, and Everything')or with the label method:
>> label(a,'NewField','Answer to the Ultimate Question of Life, the Universe, and Everything')The current label of an Alias is inquired similarly with the label method
>> label(a,'NewField')The alias labels are used as axis labels when plotting objects.
ans =
Answer to the Ultimate Question of Life, the Universe, and Everything
>> getalias(a,'NewField')In this case, that is when Aliases are defined as numerics (scalar, vector, matrix), this is strictly equivalent to
ans =
3
>> a.NewField
>> get(a,'NewField')
>> a=load(iData, [ ifitpath 'Data/sv1850.scn'])which makes it so that NewField is de-facto QH.
>> a.QH
ans =
0.9791
...
1.0214
>> setalias(a,'NewField','QH')
>> setalias(a,'NewField','Data.ZEROS') % a link to the whole ZEROS subfield structureThe AnotherNewField points to NewField, which was defined as Data.ZEROS.A1.
>> setalias(a,'NewField','Data.ZEROS.A1') % a link to the A1 value in the ZEROS subfield
>> setalias(a,'AnotherNewField', 'NewField'); % creates a link to a link to a Data part (follow me !)
>> a.AnotherNewField
ans =
-115.7500
>> a.Data.NewContent1 = rand(10); % add a new array in the Data
>> a.Data.NewContent2 = struct('a',1,'b', rand(10)); % add a new structure in the Data
>> setalias(a,'HowAreYouToday', 'Data.NewContent2');
>> setalias(a,'NewField', '[ this.Data.ZEROS.A1 this.Data.VARIA.A1 ]') % an expression which is built from two linksIn this last example, we have changed the NewField definition, which immediately affects AnotherNewField (defined above).
>> a.NewField
ans =
-115.7500 39.2600
>> a.AnotherNewField
ans =
-115.7500 39.2600
>> setalias(a,'NewField', [ 'file://' ifitpath '/Data/sv1884.scn#Data' ])The file is accessed for each alias value request.
>> a=load(iData, [ ifitpath 'Data/sv1850.scn'])We see here that the get method follows definitions recursively until a numeric value is found.
>> setalias(a,'NewField','QH')
>> get(a,'NewField') % is the same as a.NewField: return the value
ans =
0.9791
...
1.0214
>> getalias(a,'NewField') % return the definition
ans =
QH
>> a=load(iData, [ ifitpath 'Data/sv1850.scn'])The direct structure-like assignment and set method propagate the assignment value to the fully qualified link.
>> setalias(a,'NewField',3) % a static scalar
>> setalias(a,'AnotherNewField', 'NewField');
>> a.NewField
ans =
3
>> a.AnotherNewField = 4 % this stores 4 in the NewField alias !
>> a=load(iData, [ ifitpath 'Data/sv1850.scn'])
>> setalias(a,'NewField',3) % a static scalar
>> setalias(a,'AnotherNewField', 'NewField');
>> get(a,'AnotherNewField') % evaluated as NewField, then as 3
ans =
3
>> a.AnotherNewField % returns the same as get
>> a=load(iData, [ ifitpath 'Data/sv1850.scn'])whereas the similar set command removes the final target PNT but leaves the NewField Alias pointing to nothing.
>> setalias(a,'NewField','PNT') % point to PNT, which is a vector here
>> setalias(a,'NewField','') % removes NewField alias
>> rmalias(a,'NewField')
>> setalias(a,'NewField','PNT')Re-defining PNT will, at the same time, re-enable NewField.
>> set(a,'NewField','') % removes PNT, leaves NewField in error state (undefined)
>> setalias(a,'NewField','QH')That's why we strongly recommend to use setalias when you wish to control an Alias immediate definition, and set when you want to control its final value. The setalias can also define the Alias as a static numerical value, as seen previously, without risking to affect deeper object parts. Similarly, a call to getalias will reveal the nature of the Alias, as link or numerical value.
>> set(a,'NewField',1) % will set QH to 1
>> a=load(iData, [ ifitpath 'Data/sv1850.scn']) % returns an iData objectIn the iData terminology, the Signal/Monitor is the rank 0 axis (see the Axis section below)
>> a.Signal % returns the Signal value as an Alias
>> get(a,'Signal')
>> getalias(a,'Signal') % returns the Signal definition, which here points to the CNTS Alias
>> getaxis(a, 0 ) % request Signal/Monitor value as an axisSetting the axis 0 with a value first multiplies it by the Monitor, and then sets the Signal. In case you want to ignore the Monitor weighting, simply assign the Signal directly.
>> a{0}
>> label(a,'Signal') % returns the current Signal labelThis label is shown on plots along the Signal axis (e.g. Y on 1D plots and Z on 2D plots). We point out that the object can be assigned a global Title and Label as well. The title is shown as the plot title and the object text display (see Inquiring the iData object).
>> label(a,'Signal', 'Make it so ?')
>> label(a, 0, , 'Make it so ?')
>> a=load(iData, [ ifitpath 'Data/sv1850.scn'])creates a new iData object with a subscript Signal (and relevant axes definitions - see below).
>> a([ 2 4 6 8 9])
>> a=load(iData, [ ifitpath 'Data/sv1850.scn'])In principle, there is no limitation concerning the size of the Data and Signal, and the dimensionality (except memory restrictions). The following table indicates the different types of Signal that the iData can handle, and the associated requirements regarding Axes.
>> ndims(a)
ans =
1
>> size(a)
ans =
15 1
| Dimensionality |
Signal size |
Related axes |
| empty ndims=0 |
Signal is empty |
No axis |
| 0D scalar ndims=1 |
Signal is a single number |
In principle, no axis is
needed here. |
| 1D vector ndims=1 |
Signal is a vector |
One Axis required, as
scalar or vector |
| 2D matrix ndims=2 |
Signal is a matrix |
Two axes required, as
scalar, vector or matrix matching Signal |
| 3D volume ndims=3 |
Signal is a volume |
Three Axes are required, as scalar, vector, matrix or volume matching Signal |
| nD grid space ndims=n |
Signal is nD |
n Axes are required, each may have a
dimensionality from 1 to n, with preference for vectors. |
| nD event - column based
ndims=n |
Signal is a long vector
(1D) |
n Axes are required, each being a scalar
or vector of same length as Signal. NOTE: Not all Math
methods support this type of data, and it should be
converted, when appropriate, into nD grid histogramed with
the hist or interp operators
(see Math/interpolation). |
>> a=load(iData, [ ifitpath 'Data/sv1850.scn'])The Error defaults to sqrt(Signal) when not set (that is a.Error=[]). A constant Error can be set, an expression (including using Data or this) or any numerical value that matches the Signal size.
>> a.Signal
>> a.Error
>> getalias(a,'Signal')
ans =
CNTS
>> getalias(a,'Error')
ans =
sqrt(this.Signal)
>> a=load(iData, [ ifitpath 'Data/sv1850.scn'])
>> a.Signal
>> a.Error
>> a=load(iData, [ ifitpath 'Data/sv1850.scn'])
>> getaxis(a, 'Signal') % or: a{0}
>> getaxis(a, 'Error')
>> a=load(iData, [ ifitpath 'Data/sv1850.scn']) % returns an iData objectwhere the first command returns the 1st axis values, and the second commands returns its definition, which may be a link to a Data part.
>> getaxis(a, 1 ) % request axis 1 values, that is 'Y' along Signal rows
>> getaxis(a,'1') % request axis 1 definition
>> a{1} % returns the 1st axis values = getaxis(a,1)
>> a{'1'} % returns axis definition = getaxis(a,'1')
ans =
QH
In principle, the axis rank should be within 1 up to the object
dimensionality.>> label(a,1) % returns the 1st axis label, which is QH here
ans =
QH axis Ki=1.48
>> a=load(iData, [ ifitpath 'Data/sv1850.scn']) % returns an iData objectThe fast notation a{n} can also be used for direct assignments
>> setaxis(a, 1, 23) % directly fixes the value of the axis to a constant 23
>> setaxis(a, '1', 'QH') % defines axis 1 as 'QH', which is an alias of the object
>> a{1} = 23 % directly fixes the value of the axis to a constant 23
>> a{'1'} ='QH' % re-defines axis 1 as 'QH', which is an alias of the object
In case a new Axis
is defined with a numerical value (scalar, vector, matrix,
...), an Alias is
automatically created, and the Axis then points to it.>> label(a, 1, 'change label of QH')and there is a fast notation for axes of rank 1 to 4, similar to the Matlab Graphics Object syntax, where the 'X' and 'Y' are swapped for ranks 1 and 2 (see note above):
>> xlabel(a, '2st axis label') % except for 1d objects where it points to the first axisWARNING: when assigning an axis, in the case it has been previously defined as a link, the target of the link is assigned (refer to the relevant Alias section for more information).
>> ylabel(a, '1st axis label')
>> zlabel(a, '3st Z axis label')
>> clabel(a, '4st C axis label')
>> a=zeros(iData, size) % creates a [size] array of empty objectsThe squeeze method acts just like the usual Matlab squeeze one, and removes singleton dimensions in multi-dimensional arrays. This is useful when working with e.g. cat, dog, and array subscripts such as a(1,:,:).
>> a=zeros(object, size) % duplicates 'object' as a [size] array
>> b = [ a 2*a ];
Some data sets contain lists of 'events', that is a set of coordinates for a set of signal values. These events are then collections of 'rows', which each has structure:
[ x y z ... sx, sy, sz ...]
where x,y,z,... are the coordinates, and sx,sy,sz,... are signal values (for instance a magnetic field vector at given location in space).
On import, these data sets result in a matrix which rows are events, and columns are the coordinate/values, as above.
It is possible to format the data set into a formal event list, using the 'event' method after loading. The 'event' method can either make a guess on the dimensionality of space and signal, or specify the number of columns which are affected to the coordinates x,y,z,... (dimensionality of space), and the columns which is to be used for the 'signal'. When more that one 'signal' column is given, the norm of the given columns, per event, is used.
Event data sets can further be converted into histograms using the 'hist' or 'meshgrid' methods. The histograms can be prettified using the 'fill' method, which makes sure that empty spaces in the histogram are interpolated with neighboring values. More information available in the Math page.
>> methods iData
We shall present in particular a few additional methods of general
use that may be useful.