iFit: a simple library to analyze data
E. Farhi,
ILL/DS/CS - Version 1.10
- Aug. 22, 2017
Welcome to
the iFit Data Analysis library !
<
ifit.mccode.org>
"Simple methods to be used for
complex data analysis"
Main functionalities are
[
iData Load Plot Math Fit Treat Save Optimization
iFunc Models File formats]
[
miFit PyFit
Phonons ResLibCal
]
Purpose
The
iFit
library (pronounce [
eye-fit])
is a set of methods to load, analyze, plot, fit and optimize
models, and export results.
iFit
is based on
Matlab, but
can also be launched without Matlab license (stand-alone
version).

The
library focuses on
doing the
math right. Any text file can be imported straight
away, and a large set of binary files are supported. Any data
dimensionality can be handled, including event based data sets
(even though not all methods do work for these). Any model can
be assembled for fitting data sets.
iFit can also be used transparently from
Python /
NumPy. Refer to our
PyFit documentation.


The spirit of the library is to include object definitions, with
a set of methods that provide all the means to perform the usual
data analysis procedures.
- The iData object holds any data
set
- The iFunc object holds any
mathematical model function
The library can be used from user interfaces, or from the prompt
as a set of commands and scripts.
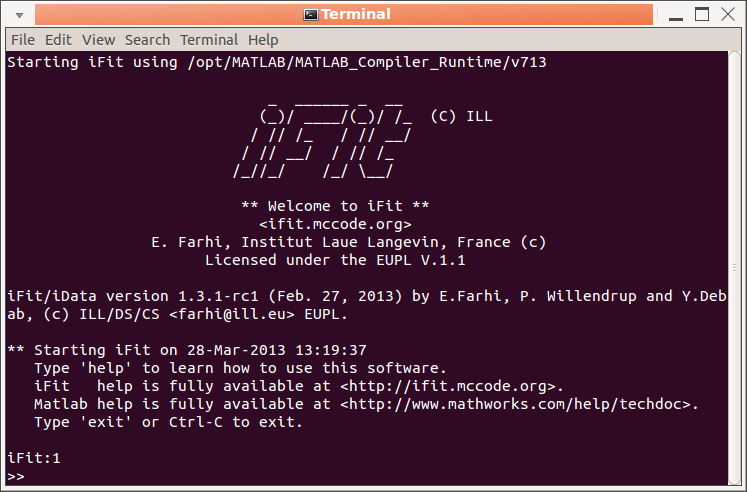
From the prompt (command
line)
|
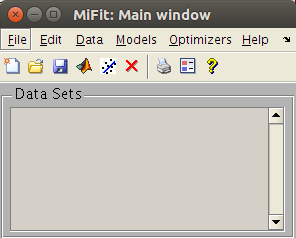
User interface: miFit
|
neutron TAS: ResLibCal |
Have a look at the Quick
Start or the iFit standalone
page. There, you mostly manipulate iData
and iFunc objects.
|
an advanced yet simple user interface to load data sets,
plot, transform, export, and fit.
|
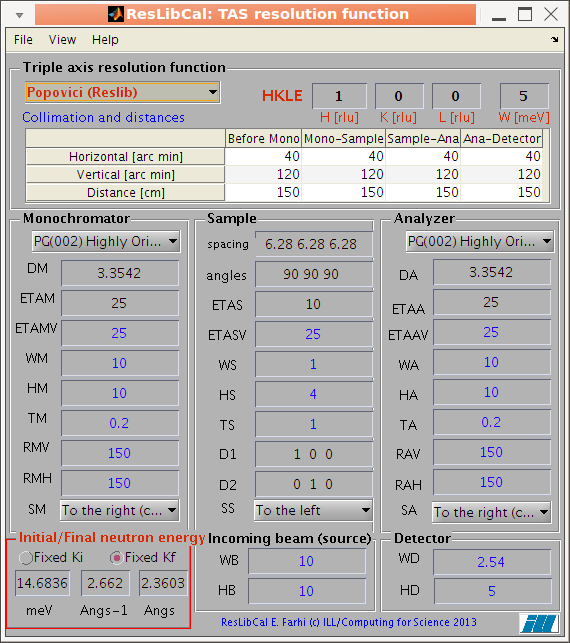
compute the neutron TAS spectrometer resolution
ellipsoid (ResCal/ResLib merge)
|
 A gallery of some of the things iFit can do
A gallery of some of the things iFit can do
Obtaining the
package
To install the package, refer to the
Install
page.
You need
Matlab to make
use of the source package. The stand-alone (binary) package does
not require Matlab, but you will need the Matlab Compiler
Runtime (refer to the
Install page). The
Python interface requires either Matlab
or the iFit standalone package to be installed, as well as
Python
and
NumPy.
The package is currently available from the
ifit.mccode.org web site:
- standalone
packages (no need for a Matlab license) for
Linux, Windows and Mac OS X are available from the Install
page. You will then use the iFit standalone command line and its miFit graphical user interface to
the package (a terminal/command window providing Matlab/iFit
capabilities).
- ISO distribution: ILL/CS
Live DVD ready-to-run system with Phonons, McStas and iFit
(standalone) all pre-installed.
Please register to the
ifit-users
mailing list.
What's in the
box...
What you will handle mainly deals with
iData
and
iFunc objects. The
miFit user interface is a simplified
entry point to these objects and their capabilities. Please
start to read these pages first, as well as the
Quick Start page if you're in a
hurry.

The miFit User interface: the iFit
main window
miFit is the main entry
point for an easy access to most iFit functionalities: import,
plot, fit, treat, export data sets and Models.
This is definitely where you should start.
The iData objects (iFit/@iData): the core data set
The
iData
objects have been defined in order to perform the following
operations seamlessly:
- Load any text formatted file, and a
large set of binary format files
- Display object information, including metadata from the
original file
- Plot the object Signal in a simple
view for 1D, 2D and 3D objects
- Perform mathematical operations using
objects (=-/* trigonometric projections integration
re-binning...)
- Fit a parametrized model to the object
Signal, which makes use of the Optimizers
and Models libraries.
- Save the object into a set of output
formats
An extended list of iData methods is
available in the
Methods page. In
addition, a few dedicated applications of these methods is
presented in the
Treatment page, for
instance for neutron scattering.
The iFunc objects (iFit/@iFunc): the core model
The iFunc objects hold models
(mathematical expressions that return a vector, matrix, ...)
value, as a function of a parameter set, and axes. You can
easily create new models, or use those from the Models page, and combine them to build
more complex models. The objects support most usual mathematical
operators.
The fit
functions/models (iFit/Models)
A predefined set of parametrized models is given in the
Models part of iFit in order to fit some
imported data and find best parameter sets which describe it.
These functions may be used by the
fits
method. All these models are
iFunc
objects. There are very advanced Models to compute for instance
4D phonon dispersions
using
ab-initio codes, neutron Monte-carlo instruments (
McCode) and other simpler analytical
methods.
The lower
level optimization library (iFit/Optimizers)
A set of optimization routines has been gathered in the
Optimizers part of iFit. It enables
to choose and compare how different optimization methods can
solve a problem. This library is used transparently by the iData
fits method, but it may also be used
independently.
The lower level import routines (iFit/Loaders)
The file import method used by iData
is performed on a lower level by the
Loaders
part of iFit. Its purpose is to import any binary or text file,
without knowing its format. A number of post-import wrappers exist
in order to reshape the imported data to match specific needs.
These wrappers are very short to write.
If needed, other importation methods should be added there, to be
able to handle more formats. This library is used transparently by
the iData
load method, but it may also be
used independently to produce simple structures describing the
data files (even though we do not recommend this).
The
documentation (iFit/Docs)
The documentation is given as web pages, and all methods/functions
have their own embedded help. To access it use:
>> doc(iData)
A list of all available
iData methods
is also available, and can be directly accessed with
>> methods(iData)
>> methods(iFunc) % equivalently for models
The documentation currently contains the following pages:
- Changes: Version releases, Changes
and Bugs list.
- Credits: Credits, Licensing and
contributions from others.
- Fit: how to fit data sets onto
mathematical models and find the best parameter set
- iData: the description of the
fundamental data set object used in iFit, properties and how
to handle it
- iFunc: the description of the
fundamental model object used in iFit, properties and how to
handle it
- Loaders: how to import data files
into structures, which can be converted into iData afterwards.
This is a sub-library which you should only use if you need
the data NOT to be an object (but you then loose most of the
iData capabilities).
- Models: model functions that cab
be use for data fitting. A function maker tool is also
provided. Specialized models provide phonon dispersions,
Rietveld refinement, small angle scattering.
- index: This iFit Welcome page.
- iFit: the interface used in the
stand-alone distribution.
- Install: the download and installation page.
- Optimizers: the low level
optimization methods.
- Load: how to load data set directly
as iData objects. This is what you should use.
- Math: all mathematical operations
and related data handling to apply on iData objects.
- McStas: an interface with the McStas neutron scattering
ray-tracing simulation code for simulation, scanning and
optimization.
- Methods: a list of all available
methods for iData objects.
- miFit: the iFit main user interface.
- Models_Phonons: a tutorial for those wishing to compute phonons in materials (lattice dynamics).
- Plot: many ways to plot the iData
objects.
- PyFit: an interface which exposes
transparently all iFit methods and objects into Python/NumPy.
- QuickStart: where you should
start (a quick tutorial).
- ResLibCal: the neutron triple-axis resolution computation tool, with a user interface.
- Save: Saving iData objects into many
file formats.
- Treatment: a set of
methodologies applied to specific cases, for instance neutron
scattering (dynamic structure factors).
- Technical documentation
You may also search this whole documentation (using Google):
Example data
files (iFit/Data)
Some
Data files are given as examples to be
used along the documentation web pages and in the
Quick Start.
Interfaces with
other software and Graphical front-ends (iFit/GUI)
Specific interfaces have been written in order to use iFit
transparently for end-user applications:
- miFit: an advanced yet simple user interface to load data sets, plot, transform, export, and fit.
- McStas: a wrapper to run
simulations or optimize McStas
and McXtrace
instruments.
- ResLibCal:
compute the neutron TAS spectrometer resolution ellipsoid
(ResCal/ResLib merge).
- Sqw_Phonons:
Phonon dispersion from ab-initio force estimate and
dynamical matrix. Have a look as well to the Models_Phonons page.
- iFunc/Model Builder:
a dialogue to create iFunc fit
function/models.
- iFit standalone: the prompt
interface used in the no-Matlab-license release. Can execute
any Matlab expression, scripts, handle iData and iFunc
objects, optimizers, plotting, ...
- PyFit: using iFit transparently
from Python/NumPy.
Ok, let's start !
To download and install the package, refer to the
Install page.
We then recommend that you start with the
miFit user interface.
If you want a quick example with commands, refer to the
Quick Start page. Otherwise, the
typical usage would be:
Start Matlab... or iFit stand-alone
>> addpath(genpath('/some/path/to/iFit')); % not needed for the stand-alone version
>> a = iData([ ifitpath 'Data/ILL_IN6.dat' ]);
>> plot(log10(a))
>> b=2*a+log(a)
>> plot(gauss2d)
>> parameters=fits(a,gauss2d)
>> saveas(a,'filename.pdf')
For beginners
If you do not know Matlab already,
you will discover that using the iFit library does not require
advanced IT knowledge. The general syntax are rather intuitive,
and all methods are able to tolerate (and correct) most of the
user mistakes. In this respect, when some of the input parameters
are missing, sensible choices can be made automatically. You can
look at the
iFit standalone page, where a short
Matlab introduction is available.
If you have
used in the past the MFit/MView
suite, you will find that this library has the same scope, but
omitting the user interface.
If you have
used the Spec1d
object, you will quickly find that the
iFit follows exactly the same
syntax.
If you come
from the Python world,
you will find out that the iFit syntax is pretty close. We provide
for you a Python interface to iFit which exposes all iFit objects
and methods into Python. Refer to the
PyFit
page.
Help me !
An extensive documentation is provided as web pages (such as this
one) which you access from Matlab with either
>> doc(iData)
>> doc(iFunc)
>> methods(iData)
and help pages for each function/method, which you can access with
e.g.
>> help iData
>> help iData/load
>> help iFunc
A Tutorial (Jan 2012)
is available as PDF presentations with exercises.
You may also search this whole documentation (using Google):
Finally, the whole Matlab documentation is available on-line for
free, especially to complement the standalone limited help system.
A list of bugs and release
history is recorded in the Changes
page.
If you find new bugs please send them to me [farhi (at) ill.fr] with:
- your Matlab version
- the iFit version which you can get from the Matlab command version(iData)
- the script/command that reproduces the error together with any
required external data file
- a copy of the error messages produced by the script/command.
- a smile ;-)
The ifit-users
is the list set-up in order to exchange information (issues,
questions, announcements). Messages should be sent to ifit-users@mccode.org.
You may register by entering your email address and press subscribe:
Credits, Links and
Disclaimer
iFit was invented by E.
Farhi and took many years to come to you.
Substantial contributions from Y.
Debab and P. Willendrup.
If you produce scientific output using iFit, I'd appreciate if you
cite the following reference:
- E. Farhi, Y. Debab and P. Willendrup, J. Neut. Res., 17 (2013) 5. DOI:
10.3233/JNR-130001
- E. Farhi, The iFit data
analysis library, ILL, Computing for Science
(2011-2012) <http://ifit.mccode.org>.
iFit is a product from the Computing
for Science group at the Institut
Laue-Langevin, Grenoble, France.
See the Credits page for more
information on contributions, including licensing.
A set of links to related
packages is also included there.
If you wish to contribute, send
suggestions, scripts, and new functions/methods to [farhi (at) ill.fr]. Your
contribution will then be included in the next release, with proper
credits to the authors.
An other solution is to use the GitHub 'pull request' mechanism,
i.e. login at GitHub, fork
your own iFit repository, modify it at your convenience, and
then mark
changes for inclusion back into the master iFit branch. These
changes will then eventually be validated, and inserted into the
project.
Disclaimer:
This software is experimental, and should not be considered
bullet-proof. In particular, expect many bugs - which should be
reported to me [farhi (at) ill.fr]
if you want them to be fixed quickly. Also, this prototype is known
to be slow. This will be improved gradually...